[without library] Iris Flower Species Classification using Gaussian Naive Bayes
[without library] Iris Flower Species Classification using Gaussian Naive Bayes
Based on - [2010] Generative and Discriminative Classifiers : Naive Bayes and Logistic Regression - Tom Mitchell
Introduction
This notebook implements Gaussian Naive Bayes. It performs multi-class classification on Iris Flower Species dataset consisting of four attributes and belonging to three classes. There are 300 data points that we divide into training and test data. The prediction accuracy is 94-100%.
Resources:
- CMU Qatar Lecture Notes - Naive Bayes - Gianni A. Di Caro
- CMU Machine Learning 10-701 - Pradeep Ravikumar
- CMU Machine Learning 10-701 - Tom Mitchell
- How is Naive Bayes a Linear Classifier? - stats.stackexchange
- Naive Bayes classifier - Wikipedia
Towards the end, I have included a sidebar on the comparison of Naive Bayes and Logistic Regression.
Taxonomy and Notes
Probabilistic, Generative
Naive Bayes is a probabilistic classifier. It learns the underlying joint distribution P(X, Y) by learning the likelihood distribution P(X\|Y) (a.k.a class-conditonal) and the prior distribution P(Y) (a.k.a class-prior). The distribution learnt can be used to generate data. Hence, it is a generative classifier. Naive bayes does not model the class decision boundaries, but instead models the distribution of the observed data. In Generative Modeling, we learn a full probabilistic model, the joint distribution P(X, Y). We assume a parametric form of the underlying probability distribution, and estimate those parameters from the observed data.
Parametric
Naive Bayes is parametric since the distribution assumed to model the likelihood (say, gaussian, multinomial, or bernoulli) have parameters. Gaussian distribution has parameters for mean and standard deviation.
Linear
In general, the Naive Bayes classifier is not linear. But, it becomes linear, if the likelihood is exponential. This is because the log-likelihood becomes linear in the log-space. Gaussian, Multinomial, or Bernoulli distributions are all from the exponential family of distributions, so Naive Bayes can be applied to data exhibiting these distributions. Under the assumption of an exponential distribution, Bernoulli Naive Bayes maps to Binomial Logistic Regression, and Multinomial Naive Bayes maps to Multinomial Logistic Regression.
Event Model
The assumptions on distributions of features are called the event model of the Naive Bayes classifier.
Event Model - Continuous Data (MNIST Digit Recognition, Iris Flower Species Classification)
When dealing with continuous data, a typical assumption is that the continuous values associated with each class are distributed according to a normal (or Gaussian) distribution. The Iris Flower Species dataset has attributes that exhibit gaussian distribution.
When assuming Gaussian class-conditionals, if all class-conditional gaussians have the same covariance, then the quadratic terms cancel out and we are left with a linear form. To take an example of MNIST dataset, if we asume that the variance across digits is the same, then we have a linear model.
The class prior distribution has to be a discrete distribution (say, multinoulli) that distributes the probability of a data point belonging to class among K possible classes.
Note: Sometimes the distribution of class-conditional marginal densities is far from normal. In these cases, kernel density estimation can be used for a more realistic estimate of the marginal densities of each class.
Event Model - Discrete Data (Text Classification)
For discrete features (document classification), Multinomial and Bernoulli distributions are popular. With a multinomial event model, features represent frequencies of events (say, count of word occurrences). With a bernoulli event model, features represent presence or absence of events (say, presence or absence of words).
Binary and Multiclass
Naive Bayes can be both binary and multiclass.
Summary
In summary, Naive Bayes is:
- Probabilistic
- Generative
- Binary and Multiclass
- Linear
- Parametric
Why Naive?
Naive Bayes makes an assumption that the input attributes are independent of each other. This results in a significant reduction in the number of parameters the model needs to learn. This is because, since each attribute is assumed to be influenced only by the class its data point belongs to, the model only has P(X_i|Y_k) terms, and no P(X_i|X_j), P(X_i|X_j, X_k), etc terms. This is the reason the model is called ‘Naive’ because it is seldom the case that the input attributes do not influence each other. Still, Naive Bayes has proven to be effective.
Due to the modeling assumptions of attributes being independent, Naive Bayes model introduces a lot more inductive bias as compared to Logistic Regression. This also results in faster convergence of order O(log N) (N is the number of data points). The fast convergence can perhaps be on less accurate estimates.
Algebraically solved Closed-Form
Naive Bayes class-conditional probabilities (maximum likelihood estimation) can be deduced analytically. Also, the class-priors can be deduced using frequentist methods. So, there is a closed-form solution to Naive Bayes. And, hence, we do not need a numerical method like gradient descent.
Imports
import numpy as np
import pandas as pd #input
from numpy.random import rand
from numpy import mean, std #mean and standard deviation for gaussian probabilities
from scipy.stats import norm #gaussian probabilities
from math import log # to calculate posterior probability
import seaborn as sns #plotting
import matplotlib.pyplot as plt #plotting
%matplotlib inline
Data
Data Configuration
class_colname = 'Species'
Iris Flower Species
f_data = '../input/iris-species/Iris.csv'
f_cols = ['SepalLengthCm', 'SepalWidthCm', 'PetalLengthCm', 'PetalWidthCm', 'Species']
Read
read the csv file
#read the csv file
df = pd.read_csv(f_data)
drop unwanted columns
#drop unwanted columns
drop_cols = list(set(df.columns) - set(f_cols))
df = df.drop(drop_cols, axis = 1)
rename last column that supposedly has a class/label
#rename the last column to 'class'
cols = df.columns.to_list()
cols[len(cols)-1] = class_colname
df.columns = cols
sanity check for data getting loaded
df.sample(5)
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species | |
|---|---|---|---|---|---|
| 44 | 5.1 | 3.8 | 1.9 | 0.4 | Iris-setosa |
| 87 | 6.3 | 2.3 | 4.4 | 1.3 | Iris-versicolor |
| 72 | 6.3 | 2.5 | 4.9 | 1.5 | Iris-versicolor |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| 76 | 6.8 | 2.8 | 4.8 | 1.4 | Iris-versicolor |
visualize
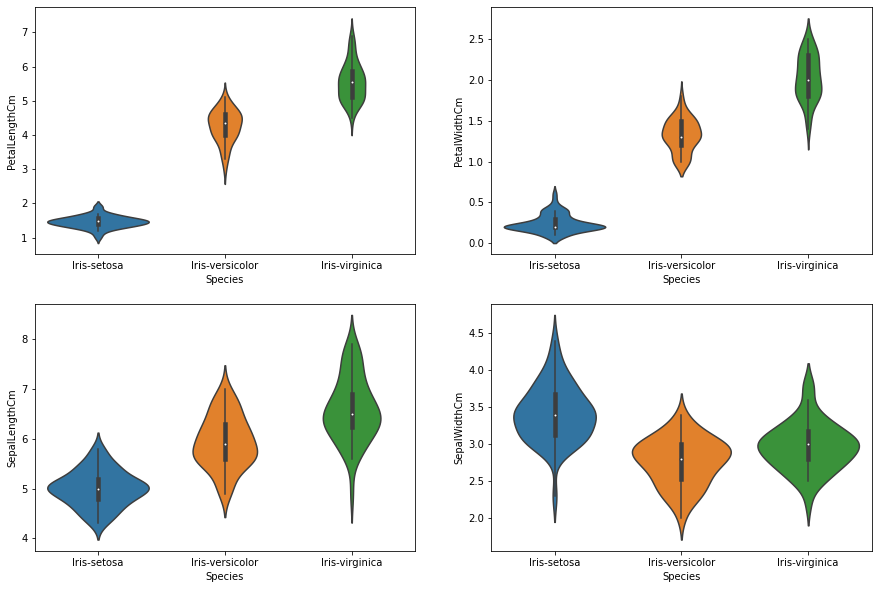
Note that the attributes exhibit gaussian distribution.
def plot_features_violin(data):
plt.figure(figsize=(15,10))
plt.subplot(2,2,1)
sns.violinplot(data=data, x='Species',y='PetalLengthCm')
plt.subplot(2,2,2)
sns.violinplot(data=data, x='Species',y='PetalWidthCm')
plt.subplot(2,2,3)
sns.violinplot(data=data, x='Species',y='SepalLengthCm')
plt.subplot(2,2,4)
sns.violinplot(data=data, x='Species',y='SepalWidthCm')
plot_features_violin(df)
Model
Model Configuration
train_ds_percent = 0.8
Training Algorithm
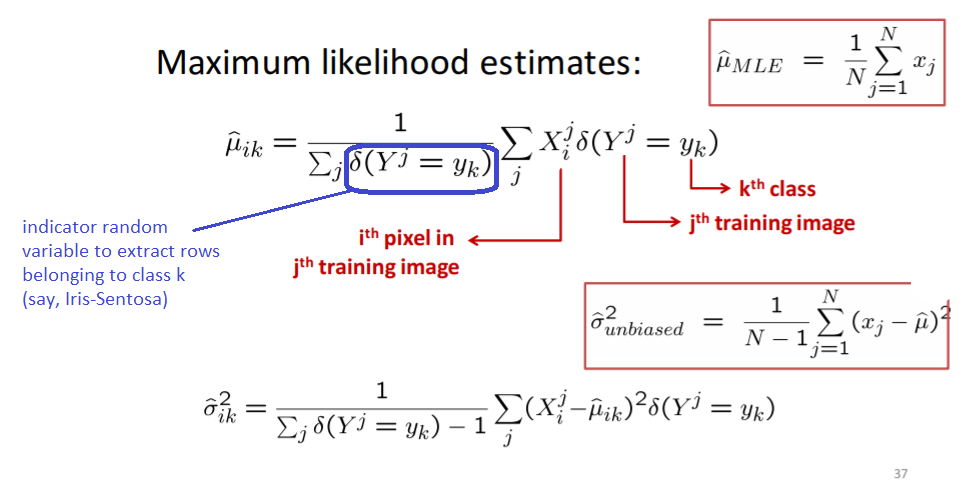
'''
return
classes: (list) of unique class names in the dataset,
got from the last column named class_colname.
features: (list) of features (column names) in the dataset.
this excludes the last column which we expect it to have the class labels.
prior: (1-d array) of dim num_classes
(prior probability of a set of features belonging to a class)
mean_std: (3-d array) of dim num_classes x num_features x 2 (2: mean and std)
(mean and standard deviation for all features, given the class)
arguments:
df: (dataframe) with features and class names (should have a 'class' column in addition to the feature columns).
class_colname: (string) provide suitable column name otherwise, using the class_colname argument.
'''
def train_gaussian_nb(df, class_colname='class'):
#number of classes
classes = df[class_colname].unique()
num_classes = len(df[class_colname].unique())
#number of features
features = df.columns[:-1]
num_features = len(features)
#number of data points
N = len(df)
#data structures for priors and
# (mean, standard deviation) pairs for each feature and class
# to later calculate likelihood (conditional probability of feature given class)
prior = np.zeros(num_classes)
mean_std = np.zeros((num_classes, num_features, 2), dtype=float)
#for each class...
for cls in range(num_classes):
#calculate prior probability of data point belonging to class cls
prior[cls] = len(df[df[class_colname]==classes[cls]]) / N
#to later calculate likelihood: conditional probability for all features, given class cls,
#we store the mean and standard deviation of all features, given class cls
for i_feature in range(num_features):
#store mean for i_feature, given cls
mean_std[cls][i_feature][0] = mean(df[df[class_colname]==classes[cls]].iloc[:, i_feature])
#store standard deviation for i_feature, given cls
mean_std[cls][i_feature][1] = std(df[df[class_colname]==classes[cls]].iloc[:, i_feature])
return classes, features, prior, mean_std
Prediction Algorithm
'''
return (integer) the (0-based) index of class to which the document belongs
arguments:
num_classes: (int) number of classes
num_features: (int) number of features
prior: (1-d array) of dim num_classes
(prior probability of a set of features belonging to a class)
mean_std: (3-d array) of dim num_classes x num_features x 2 (2: mean and std)
(mean and standard deviation for all features, given the class)
x: (list) of features
'''
def apply_gaussian_naive_bayes(num_classes, num_features, prior, mean_std, x):
#initialize score for each class to zero
score = np.zeros((num_classes), dtype=float)
#for each class...
for cls in range(num_classes):
#print('class:', cls)
#for this class, add the log-prior probability to the score
score[cls] += log(prior[cls], 10) #log to the base 10
#for each feature, add the log-likelihood to the score
for i_feature in range(num_features):
#print('feature', i_feature)
#calculate likelihood from the trained mean and standard deviation
mu = mean_std[cls][i_feature][0]
sigma = mean_std[cls][i_feature][1]
likelihood = norm(mu, sigma).pdf(x[i_feature])
#add the log-likelihood to the score
score[cls] += log(likelihood, 10) #log to the base 10
#return the index of class with the maximum-a-posterior probability
return score.argmax()
Learn
Divide data into Train and Test
#mask a % of data for training, and the remaining for testing
mask = rand(len(df)) < train_ds_percent
df_train = df[mask]
df_test = df[~mask]
Learn
#train the prior and likelihood on observed data df_train
classes, features, prior, mean_std = train_gaussian_nb(df_train, class_colname)
Prediction
Predict
#iterate over test dataset and count the number of correct and incorrect predictions
count_correct, count_incorrect = 0, 0
for index, row in df_test.iterrows():
#actual class
actual_cls = row[class_colname]
#predicted class
# input provided as row[:-1].to_list(), means, all columns except last, converted to a list
pred_cls = apply_gaussian_naive_bayes(len(classes), len(features), prior, mean_std, row[:-1].to_list())
if classes[pred_cls] == actual_cls:
count_correct += 1
else:
count_incorrect += 1
#print('(predicted, actual):', classes[pred_cls], row[class_colname])
Prediction Accuracy
print('Correct: ', count_correct, 'Incorrect: ', count_incorrect)
print('Percentage of correct predictions: ', (count_correct * 100)/(count_correct + count_incorrect))
Correct: 36 Incorrect: 2
Percentage of correct predictions: 94.73684210526316
Sidebar - Comparison of Naive Bayes and Logistic Regression
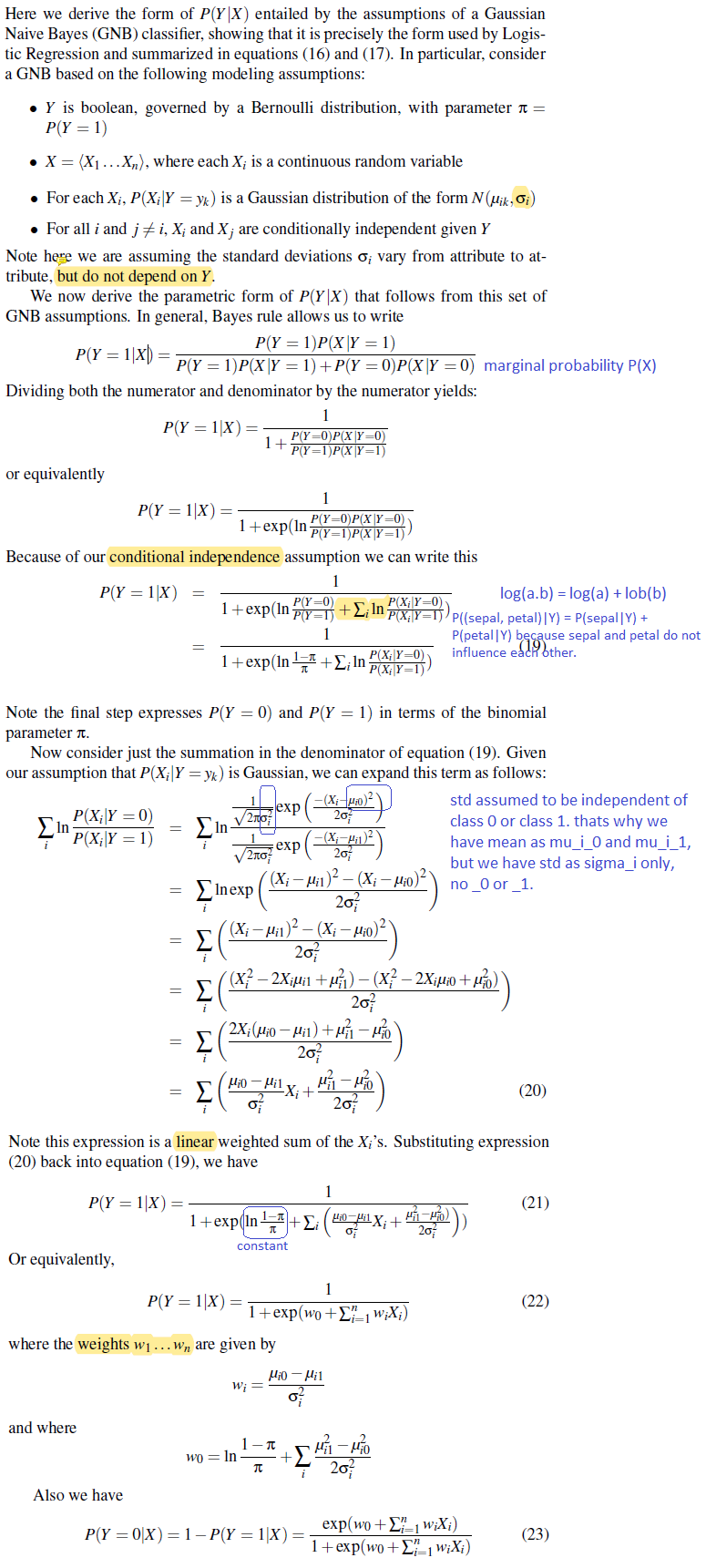
Generative and Discriminative Classifiers : Naive Bayes and Logistic Regression - Tom Mitchell
Same Parametric Form (under attribute independence assumptions)
The parametric form of P(Y|X) used by Logistic Regression is precisely the form implied by the assumptions of a Gaussian Naive Bayes clasifier.
Attribute Independence Assumption; Convergence; Input Data Size

Asymptotic Comparison
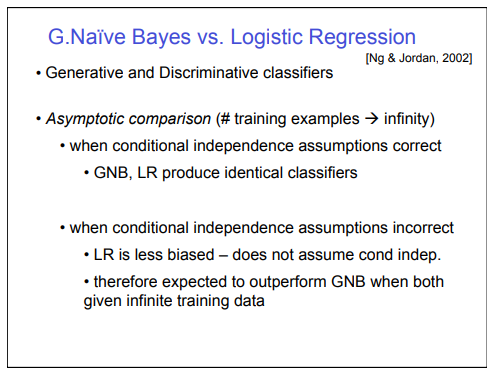
Summary